
Templates Community /
Protein Fold Evofermer Design
Protein Fold Evofermer Design
yu chungong
Published on 2022-12-28

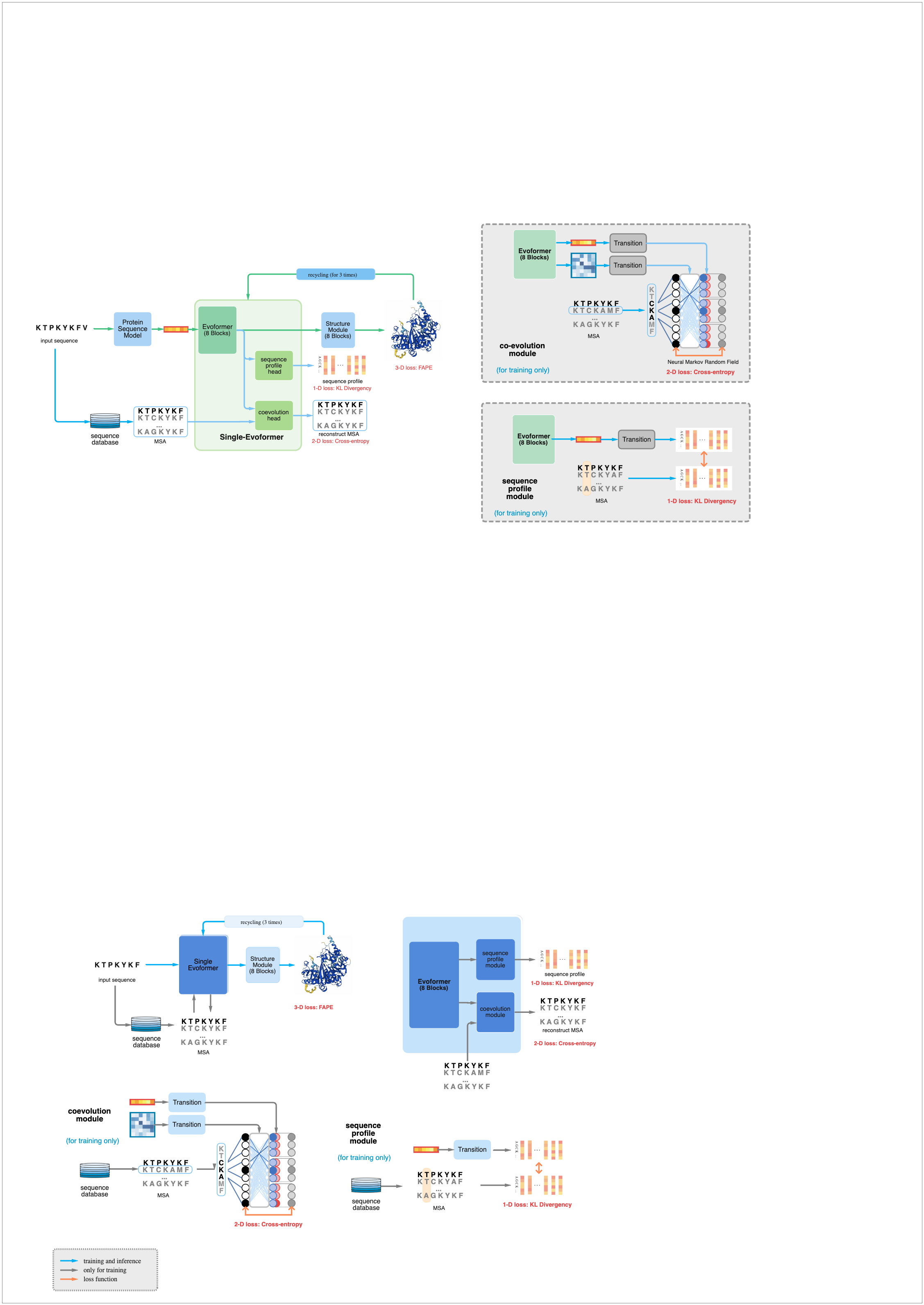
Protein Fold Evofermer is an artificial intelligence-based protein-folding and structure prediction tool developed for use in biochemistry research. It uses deep reinforcement learning to accurately predict the 3D structure of a given protein from its primary sequence of amino acids, with no prior knowledge of the target protein's three-dimensional structure. Protein Fold Evofermer can automatically recognize essential biochemical features such as active sites, secondary structures, and binding grooves for design optimization. Protein Fold Evofermer is a deep learning model developed to predict the geometric characteristics of proteins. It takes into account the C-alpha atom coordinates and defines a novel hydrogen bond potential. This model has opened up new possibilities for ab initio protein structure prediction, as it can accurately map inter-residue contacts and distances. With the help of EdrawMax, you can easily recreate a similar diagram.
Tag
IT
Protein Fold Evofermer
Share
Report
0
49

Post
Recommended Templates
Loading
